Thursday, 13 January 2022
07:58 PM
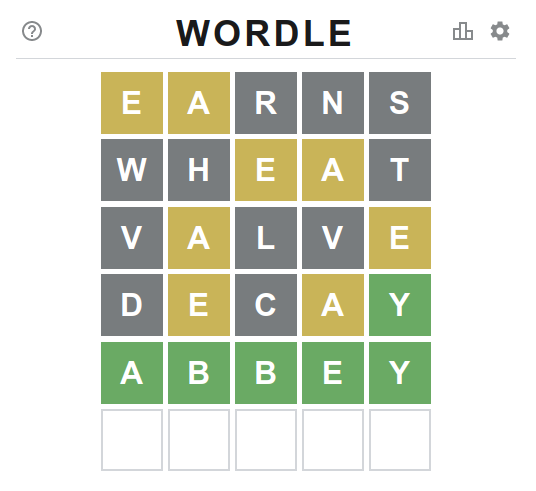
Do-It-Yourself warnings categories [blogs.perl.org] 07:58 PM, Thursday, 13 January 2022 03:20 PM, Thursday, 13 January 2022
One of the reasons I have not "moved on" from Perl to some other
more "modern" language is that Perl gives me such great access to
its inner workings. The
Do-It-Yourself Lexical Pragmas post from a couple weeks ago is
an example of this. Another example is that Perl lets you tie your
own code into its warnings
system.
Tying into the warnings machinery requires a
module. That is, the interface assumes you are reporting problems
relative to another name space that invoked your code. Your module
can either add diagnostics to existing Perl warning categories or
actually create new categories. In either case your diagnostics are
sensitive to the enablement or disablement of the category, as well
as its fatalization.
In addition to enabling or disabling warning categories,
use warnings ... and no warnings ...;
make some subroutines available which can be used to issue your own
diagnostics. These are reported relative to the file and line that
called into your module (sort of like carp()). A
useful subset of the warnings:: subroutines is:
warnings::enabled( $category )- This subroutine returns a true value if the given warnings category is enabled by the calling package, or a false value if it is not.
warnings::warn( $category, $message )- This subroutine issues the given
$messageas a warning in the given$category, or as a fatal (though trappable) error if the category has been fatalized. warnings::warnif( $category, $message )- This convenience subroutine is equivalent to
warnings::warn( $category, $message ) if warnings::enabled( $category ).
The
deprecated warning was the subject of one of my blog posts a
couple months ago. That post described its use by Perl itself, but
module authors need to deprecate code as well. You can issue your
own deprecated diagnostics like this:
warnings::warnif( deprecated => 'This feature is deprecated' );
This will be fatal if deprecated has been
fatalized, or suppressed if it has been disabled. The file and line
number appended will be those of the most-recent call into your
module, not necessarily the caller of the subroutine that contains
this code. This means you can centralize your deprecation code
without worrying about the depth of the call tree.
In order to create a new warning category named after your module, all you have to do is
use warnings::register;
Once Perl compiles this, you can treat it just like a built-in
category. If your module is named My::Module, users of
it can
use warnings 'My::Module';
or
no warnings 'My::Module';
The latter example may be more to the point, since use
warnings; enables custom categories as well.
Your module generates diagnostics in the new category using the same interface as for the built-ins. For example,
warnings::warnif( __PACKAGE__, 'Danger, Will Robinson!' );
You can actually omit the $category argument in
this case, simplifying the above to
warnings::warnif( 'Danger, Will Robinson!' );
If your module needs more than one new warning category, you can
give their base names as arguments to use
warnings::register;.
package My::Module;
use warnings::register qw{ fu bar };
will create warning categories My::Module,
My::Module::fu, and My::Module::bar.
Somewhat to my surprise, I found no documented restrictions on the names of packages that can be made into warnings categories. From the standpoint of Perl's warning machinery,
package deprecated; use warnings::register;
is not a problem, at least under Perl 5.34.0. Nevertheless, from the standpoint of both users and maintainers of such a module, this looks to me like a Very Bad Thing.
06:15 PM
Memorial [Futility Closet] 06:15 PM, Thursday, 13 January 2022 06:40 PM, Thursday, 13 January 2022

In Vienna’s Judenplatz stands a construction of steel and concrete that takes the shape of a library turned inside out. Its walls are filled with books, but the spines are all turned inward, so the knowledge they contain is inaccessible. It bears two large doors, but these do not open.
It is a memorial to the Austrian victims of the Holocaust. Artist Rachel Whiteread said, “It was clear to me from the outset that my proposal had to be simple, monumental, poetic and non-literal. I am a sculptor: not a person of words but of images and forms.”
At the unveiling, Simon Wiesenthal said, “This monument shouldn’t be beautiful. It must hurt.”
04:38 PM
[$] The first half of the 5.17 merge window [LWN.net] 04:38 PM, Thursday, 13 January 2022 04:40 PM, Thursday, 13 January 2022
As of this writing, just short of 7,000 non-merge commits have been pulled into the mainline kernel repository for the 5.17 release. The changes pulled thus far bring new features across the kernel; read on for a summary of what has been merged during the first half of the 5.17 merge window.
04:00 PM
Bits from Debian: New Debian Developers and Maintainers (November and December 2021) [Planet Debian] 04:00 PM, Thursday, 13 January 2022 04:40 PM, Thursday, 13 January 2022

The following contributors got their Debian Developer accounts in the last two months:
- Douglas Andrew Torrance (dtorrance)
- Mark Lee Garrett (lee)
The following contributors were added as Debian Maintainers in the last two months:
- Lukas Matthias Märdian
- Paulo Roberto Alves de Oliveira
- Sergio Almeida Cipriano Junior
- Julien Lamy
- Kristian Nielsen
- Jeremy Paul Arnold Sowden
- Jussi Tapio Pakkanen
- Marius Gripsgard
- Martin Budaj
- Peymaneh
- Tommi Petteri Höynälänmaa
Congratulations!
02:29 PM
Security updates for Thursday [LWN.net] 02:29 PM, Thursday, 13 January 2022 02:40 PM, Thursday, 13 January 2022
Security updates have been issued by Debian (epiphany-browser, lxml, and roundcube), Fedora (gegl04, mingw-harfbuzz, and mod_auth_mellon), openSUSE (openexr and python39-pip), Oracle (firefox and thunderbird), Red Hat (firefox and thunderbird), SUSE (apache2, openexr, python36-pip, and python39-pip), and Ubuntu (apache-log4j1.2, ghostscript, linux, linux-gcp, linux-gcp-5.4, linux-hwe-5.4, and systemd).
09:00 AM
Daniel Lange: Leveling the playing field for non-native speakers [Planet Debian] 09:00 AM, Thursday, 13 January 2022 10:00 AM, Thursday, 13 January 2022
06:14 AM
Efficiency [Futility Closet] 06:14 AM, Thursday, 13 January 2022 06:20 AM, Thursday, 13 January 2022

In the 1920s, Fiat’s car factory in Turin, Italy, contained a spiral roadway — raw materials went in at ground level, and the cars ascended as they were assembled. At the top they emerged onto a test track on the roof.
Lauded at the time, the factory was eventually outmoded and has since been remodeled into a hotel and shopping mall, but the test track remains and is open to visitors.
06:00 AM
A comprehensive comparison of supervised and unsupervised methods for cell type identification in single-cell RNA-seq [RNA-Seq Blog] 06:00 AM, Thursday, 13 January 2022 12:40 PM, Thursday, 13 January 2022
The cell type identification is among the most important tasks in single-cell RNA-sequencing (scRNA-seq) analysis. Many in silico methods have been developed and can be roughly categorized as either supervised or unsupervised. In this study, A team led by researchers at Emory University investigated the performances of 8 supervised and 10 unsupervised cell type identification methods using 14 public scRNA-seq datasets of different tissues, sequencing protocols and species. The researchers investigated the impacts of a number of factors, including total amount of cells, number of cell types, sequencing depth, batch effects, reference bias, cell population imbalance, unknown/novel cell type, and computational efficiency and scalability. Instead of merely comparing individual methods, they focused on factors’ impacts on the general category of supervised and unsupervised methods.
The workflow of the scRNAIdent pipeline
scRNAIdent provides a modularized R pipeline tool for automating the evaluation and comparison of cell typing methods in scRNA-seq analysis.
The researchers found that in most scenarios, the supervised methods outperformed the unsupervised methods, except for the identification of unknown cell types. This is particularly true when the supervised methods use a reference dataset with high informational sufficiency, low complexity and high similarity to the query dataset. However, such outperformance could be undermined by some undesired dataset properties investigated in this study, which lead to uninformative and biased reference datasets. In these scenarios, unsupervised methods could be comparable to supervised methods. This study not only explained the cell typing methods’ behaviors under different experimental settings but also provided a general guideline for the choice of method according to the scientific goal and dataset properties. Finally, the evaluation workflow is implemented as a modularized R pipeline that allows future evaluation of new methods.
Availability: All the source codes are available at https://github.com/xsun28/scRNAIdent.
05:00 AM
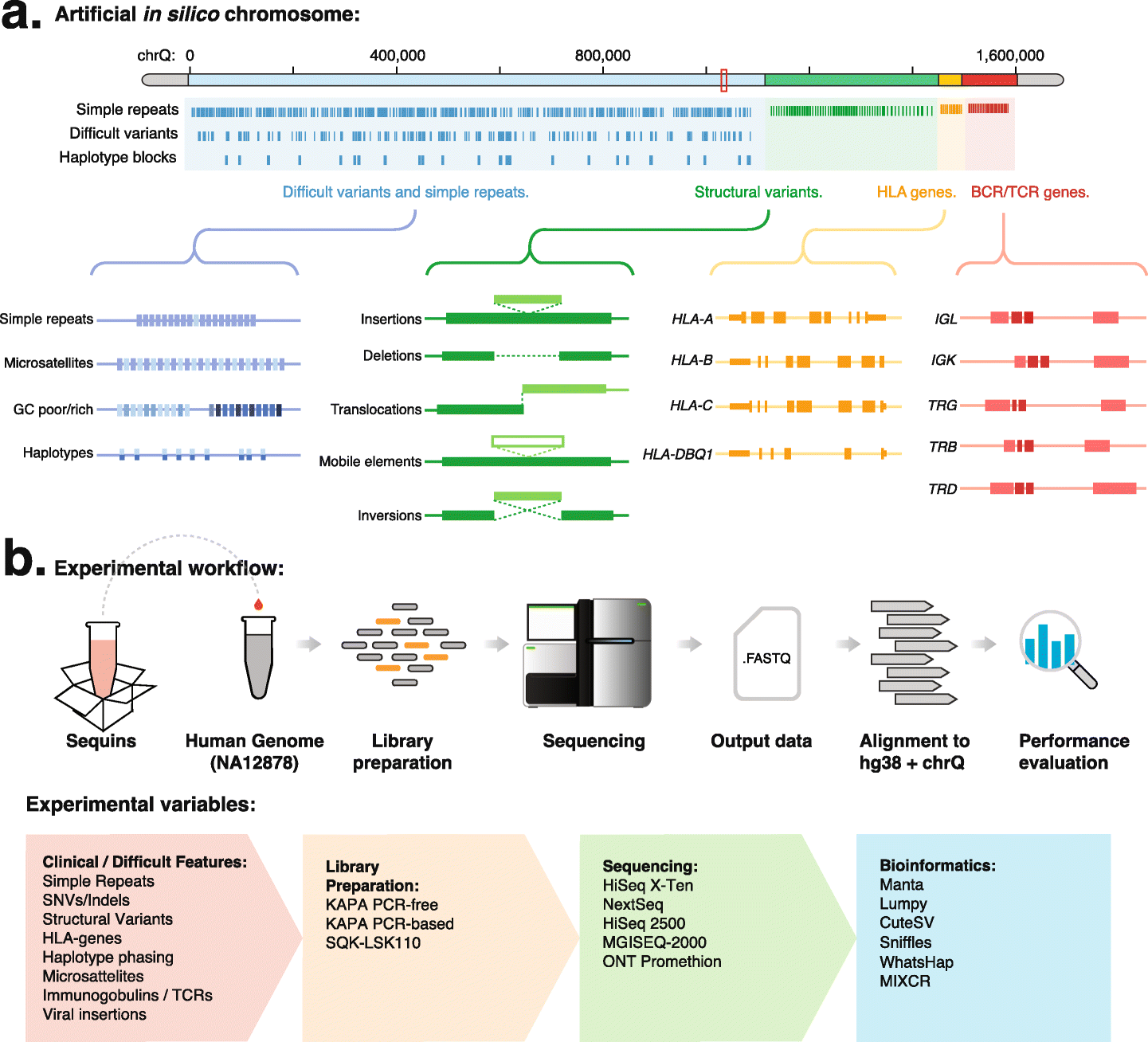
Using synthetic chromosome controls to evaluate the sequencing of difficult regions within the human genome [RNA-Seq Blog] 05:00 AM, Thursday, 13 January 2022 12:40 PM, Thursday, 13 January 2022
Next-generation sequencing (NGS) can identify mutations in the human genome that cause disease and has been widely adopted in clinical diagnosis. However, the human genome contains many polymorphic, low-complexity, and repetitive regions that are difficult to sequence and analyze. Despite their difficulty, these regions include many clinically important sequences that can inform the treatment of human diseases and improve the diagnostic yield of NGS.
To evaluate the accuracy by which these difficult regions are analyzed with NGS, a team led by researchers at the Garvan Institute of Medical Research built an in silico decoy chromosome, along with corresponding synthetic DNA reference controls, that encode difficult and clinically important human genome regions, including repeats, microsatellites, HLA genes, and immune receptors. These controls provide a known ground-truth reference against which to measure the performance of diverse sequencing technologies, reagents, and bioinformatic tools. Using this approach, the researchers provide a comprehensive evaluation of short- and long-read sequencing instruments, library preparation methods, and software tools and identify the errors and systematic bias that confound our resolution of these remaining difficult regions.
In silico chromosome design and experimental workflow
a The in silico decoy chromosome is designed to incorporate difficult and clinically important features of the human genome. The chromosome is divided into (i) small variants (including SNPs and indels) and simple repeats, (ii) structural variants (including large insertions, deletions, duplications, inversions, and translocations), (iii) HLA genes, and (iv) immune receptor genes. b The schematic diagram illustrates the use of synthetic DNA controls (sequins) and the in silico chromosome during the NGS workflow (upper panel). The range of experimental variables evaluated within this study, including difficult genetic features, library preparation methods, sequencing instruments, and bioinformatic tools, are indicated (lower panel)
This study provides an analytical validation of diagnosis using NGS in difficult regions of the human genome and highlights the challenges that remain to resolve these difficult regions.
01:06 AM
[$] LWN.net Weekly Edition for January 13, 2022 [LWN.net] 01:06 AM, Thursday, 13 January 2022 01:20 AM, Thursday, 13 January 2022
The LWN.net Weekly Edition for January 13, 2022 is available.