Wednesday, 12 January 2022
10:39 PM
[$] Relocating Fedora's RPM database [LWN.net] 10:39 PM, Wednesday, 12 January 2022 10:40 PM, Wednesday, 12 January 2022
The deadlines for various kinds of Fedora 36 change proposals have mostly passed at this point, which led to something of a flurry of postings to the distribution's devel mailing list over the last month. One of those, for a seemingly fairly innocuous relocation of the RPM database from /var to /usr, came in right at the buzzer for system-wide changes on December 29. There were, of course, other things going on around that time, holidays, vacations, and so forth, so the discussion was relatively muted until recently. Proponents have a number of reasons why they would like to see the move, but there is resistance, as well, that is due, at least in part, to the longstanding "tradition" of the location for the database.
06:24 PM
“She Was as Cute as a Washtub” [Futility Closet] 06:24 PM, Wednesday, 12 January 2022 06:40 PM, Wednesday, 12 January 2022
Raymond Chandler similes:
“He looked about as inconspicuous as a tarantula on a
slice of angel food.”
“She looked almost as hard to get as a haircut.”
“The smell of old dust hung in the air as flat and stale as a
football interview.”
“Her face fell apart like a bride’s pie
crust.”
“This car sticks out like spats at an Iowa picnic.”
“I belonged in Idle Valley like a pearl onion on a banana
split.”
“A few locks of dry white hair clung to his scalp, like wild
flowers fighting for life on a bare rock.”
“Then she straightened the bills out on the desk and put one
on top of the other and pushed them across. Very slowly, very
sadly, as if she was drowning a favorite kitten.”
“If you use similes,” he once suggested, “try and make them both extravagant and original.”
05:30 PM
Michael Prokop: Revisiting 2021 [Planet Debian] 05:30 PM, Wednesday, 12 January 2022 06:00 PM, Wednesday, 12 January 2022


Uhm yeah, so this shirt didn’t age well. :) Mainly to recall what happened, I’m once again revisiting my previous year (previous edition: 2020).
2021 was quite challenging overall. It started with four weeks of distance learning at school. Luckily at least at school things got back to "some kind of normal" afterwards. The lockdowns turned out to be an excellent opportunity for practising Geocaching though, and that’s what I started to do with my family. It’s a great way to grab some fresh air, get to know new areas, and spend time with family and friends – I plan to continue doing this. :)
We bought a family season ticket for Freibäder (open-air baths) in Graz; this turned out to be a great investment – I enjoyed the open air swimming with family, as well as going for swimming laps on my own very much, and plan to do the same in 2022. Due to the lockdowns and the pandemics, the weekly Badminton sessions sadly didn’t really take place, so I pushed towards the above-mentioned outdoor swimming and also some running; with my family we managed to do some cycling, inline skating and even practiced some boulder climbing.
For obvious reasons plenty of concerts I was looking forward didn’t take place. With my parents we at least managed to attend a concert performance of Puccinis Tosca with Jonas Kaufmann at Schloßbergbühne Kasematten/Graz, and with the kids we saw "Robin Hood" in Oper Graz and "Pippi Langstrumpf" at Studiobühne of Oper Graz. The lack of concerts and rehearsals once again and still severely impacts my playing the drums, including at HTU BigBand Graz. :-/
Grml-wise we managed to publish release 2021.07, codename JauKerl. Debian-wise we got version 11 AKA bullseye released as new stable release in August.
For 2021 I planned to and also managed to minimize buying (new) physical stuff, except for books and other reading stuff. Speaking of reading, 2021 was nice — I managed to finish more than 100 books (see “Mein Lesejahr 2021“), and I’d like to keep the reading pace.
Now let’s hope for better times in 2022!
05:27 PM
Steinar H. Gunderson: Training apps [Planet Debian] 05:27 PM, Wednesday, 12 January 2022 06:00 PM, Wednesday, 12 January 2022

I've been using various training apps (and their associated web sites) since 2010 now, forward-porting data to give me twelve years of logs. (My primary migration path has been CardioTrianer → Endomondo → Strava.) However, it strikes me that they're just becoming worse and worse, and I think I've figured out why: What I want is a training site with some social functions, but what companies are creating are social networks. Not social networks about training; just social networks.
To be a bit more concrete: I want something that's essentially a database. I want to quickly search for workouts in a given area and of a given length, and then bring up key information, compare, contrast, get proper graphs (not something where you can't see the difference between 3:00/km and 4:00/km!), and so on. (There's a long, long list of features and bugs I'd like to get fixed, but I won't list them all.)
But Strava is laser-focused on what's happened recently; there's a stream with my own workouts and my friends', just like a social network, and that's pretty much the main focus (and they have not even tried to solve the stream problem; if I have a friend that's too active, I don't see anything else). I'd need to pay a very steep price (roughly $110/year, the same price of a used GPS watch!) to even get a calendar; without that, I need to go back through a super-slow pagination UI 20 and 20 workouts at a time to even see something older.
Garmin Connect is somewhat better here; at least I can query on length and such. (Not so strange; Garmin is in the business of selling devices, not making social networks.) But it's very oriented around one specific watch brand, and it's far from perfect either. My big issue is that nobody's even trying, it seems. But I guess there's simply no money in that.
04:37 PM
IPython 8.0 released [LWN.net] 04:37 PM, Wednesday, 12 January 2022 04:40 PM, Wednesday, 12 January 2022
Version 8.0 of the IPython read-eval-print-loop implementation for Python is out.
This major release comes with many improvements to the existing codebase and several new features. These new features are code reformatting with Black in the CLI, ghost suggestions, and better tracebacks which highlight the error node, thus making complex expressions easier to debug.
03:30 PM
Malcolm: Prevent Trojan Source attacks with GCC 12 [LWN.net] 03:30 PM, Wednesday, 12 January 2022 04:00 PM, Wednesday, 12 January 2022
David Malcolm describes some GCC improvements to defend against bidirectional-text attacks in source code.
My colleague Marek Polacek and I implemented a new warning for GCC 12, -Wbidi-chars, for detecting Trojan Source attacks involving Unicode control characters. Marek implemented the guts of the warning, but when I tried it out on the examples provided by the Trojan Source researchers, I found I had trouble understanding the initial results—precisely because of the obfuscation itself.So for GCC 12, I've added a new flag to GCC diagnostics, indicating that the diagnostic itself relates to source code encoding. When any such diagnostic is printed, GCC will now escape non-ASCII characters in the source code.
03:26 PM
Security updates for Wednesday [LWN.net] 03:26 PM, Wednesday, 12 January 2022 04:00 PM, Wednesday, 12 January 2022
Security updates have been issued by Debian (cfrpki, gdal, and lighttpd), Fedora (perl-CPAN and roundcubemail), Mageia (firefox), openSUSE (jawn, kernel, and thunderbird), Oracle (kernel, openssl, and webkitgtk4), Red Hat (cpio, idm:DL1, kernel, kernel-rt, openssl, virt:av and virt-devel:av, webkit2gtk3, and webkitgtk4), Scientific Linux (openssl and webkitgtk4), SUSE (kernel and thunderbird), and Ubuntu (apache-log4j2, ghostscript, and lxml).
02:30 PM
Dramatic Improbable Readings for Arisia 2022 (local copy) [] 02:30 PM, Wednesday, 12 January 2022 10:40 PM, Monday, 17 January 2022
Improbable Research organizes a Dramatic Improbable Readings session at Arisia every year — dramatic readings from published research studies that make people laugh, then think.
In 2022, The Covid-19 pandemic led Arisia to cancel the 2022 convention. Undaunted, Improbable Research did and video recorded a new Dramatic Improbable Readings session. Marc Abrahams, editor of the Annals of Improbable Research, comperes. The dramatic readers are: Mason Porter, Sonya Taafe, Dean Grodzins, and Gary Dryfoos. Michele Liguori is the timekeeper. David Kessler is the deobfuscator.
06:28 AM
Over There [Futility Closet] 06:28 AM, Wednesday, 12 January 2022 06:40 AM, Wednesday, 12 January 2022

In 1942, homesick GI Carl K. Lindley was ordered to repair a local signpost in the Yukon. He decided to add an indicator pointing to his hometown: DANVILLE, ILL. 2835 MILES. Others began adding their own signs, and today the “Sign Post Forest” holds 80,000 signs. It’s actively accepting more — you can bring your own or make one at the visitor information center.
06:00 AM
LMU researchers have developed a method to extract more information from RNA sequencing data [RNA-Seq Blog] 06:00 AM, Wednesday, 12 January 2022 01:20 PM, Wednesday, 12 January 2022
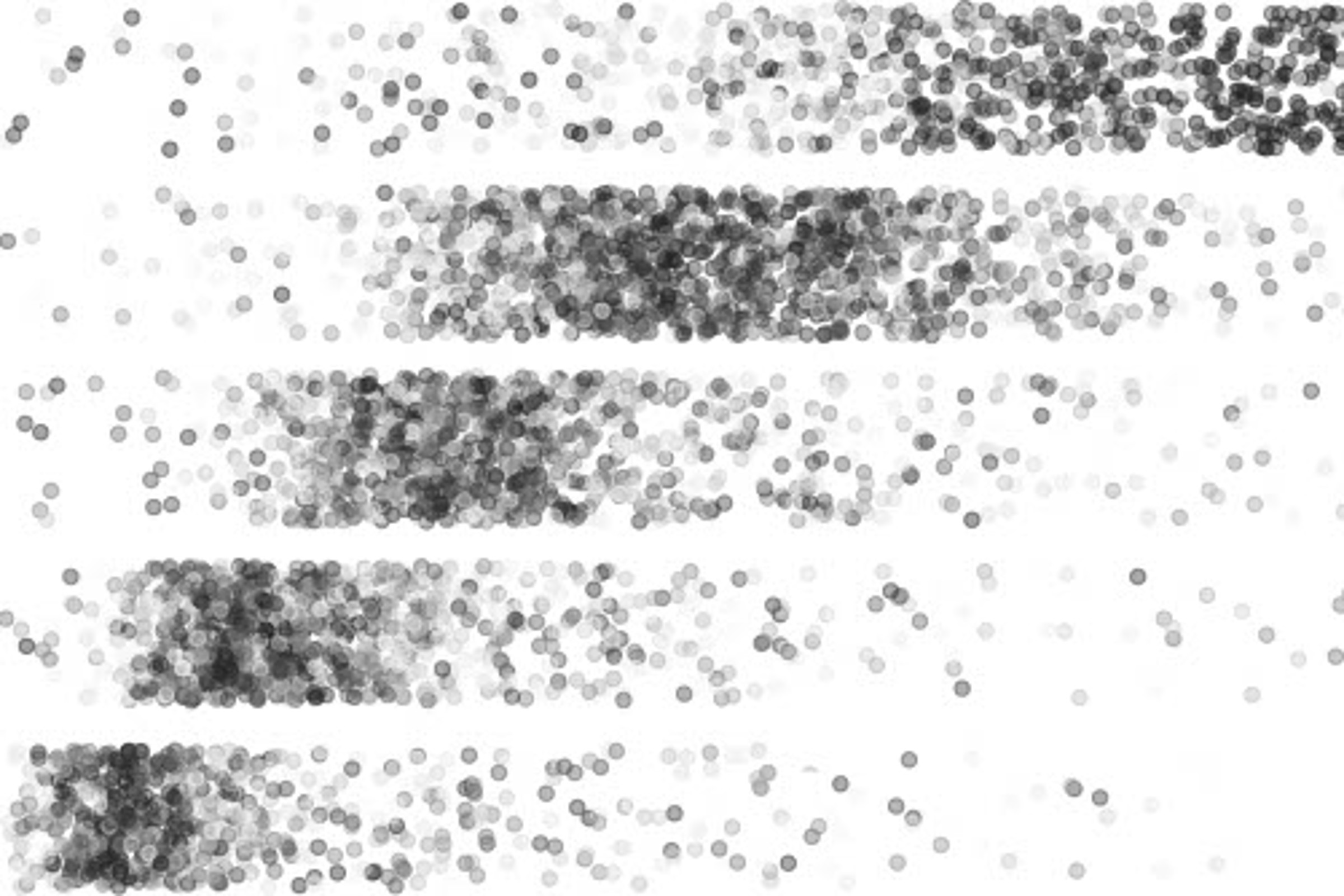
Points represent mRNA molecules that travel different distances in an electric field according to their size. Large molecules (right) are separated in Laddder-seq from small molecules (left) as along the steps in a ladder.
Higher organisms store their genetic material in the nuclei of cells as deoxyribonucleic acid (DNA). In a process called transcription, individual segments, the genes, are converted into messenger ribonucleic acids (mRNAs). Subsequently, the translation process produces proteins as the most important functional units.
Cells can turn various genes on or off. And diseases also have characteristic patterns of gene expression. This is why scientists try to sequence mRNA molecules so as to extract further information from them. The problem with established technologies is that information was often lost along the way.
A team led by Dr. Stefan Canzar from the Gene Center Munich has now presented Ladder-seq in Nature Biotechnology: a method that leads to better results. The scientists combine changes to the sequencing protocol with computer-assisted techniques. With the additional information, Canzar’s team is able, for example, to decode the function of regulatory units of neural stem cells in the brains of mice.
A jigsaw puzzle with additional information
“In sequencing before now, the first step was to break down mRNA molecules, leaving you, figuratively speaking, with the jumbled pieces of a jigsaw puzzle,” explains Canzar. Modern devices then sequence these fragments in parallel, which saves time. With powerful computers, individual portions of the sequence are put back together again. Over the past 10-15 years, several methods have been further optimized. “Nevertheless, we know that information gets lost in the process that cannot be reconstructed,” says the LMU researcher.
Dr. Francisca Rojas Ringeling and Shounak Chakraborty from Canzar’s group have amended the usual protocol. Before the actual sequencing, researchers separate RNA molecules based on their length. The easiest way to do this is a technology called electrophoresis for separating molecules in an electric field. According to their size, mRNAs travel different distances. In the gel, they appear as a ladder, hence the name Ladder-seq. Only then do the researchers carry out fragmentation and sequencing. Algorithms use information about size to piece together individual parts of the puzzle with greater precision than previously possible.
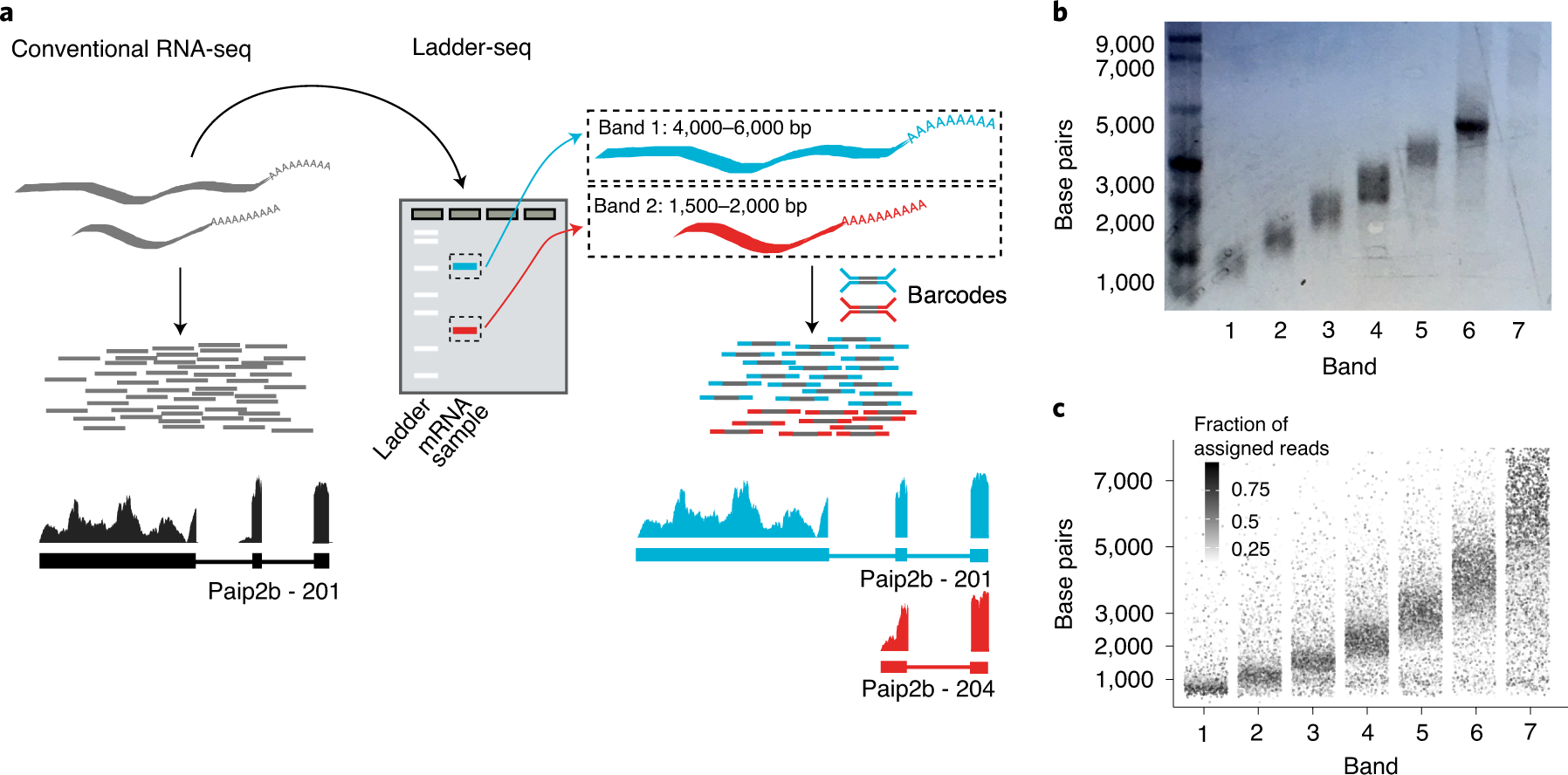
Ladder-seq uses mRNA length information to aid transcriptome reconstruction
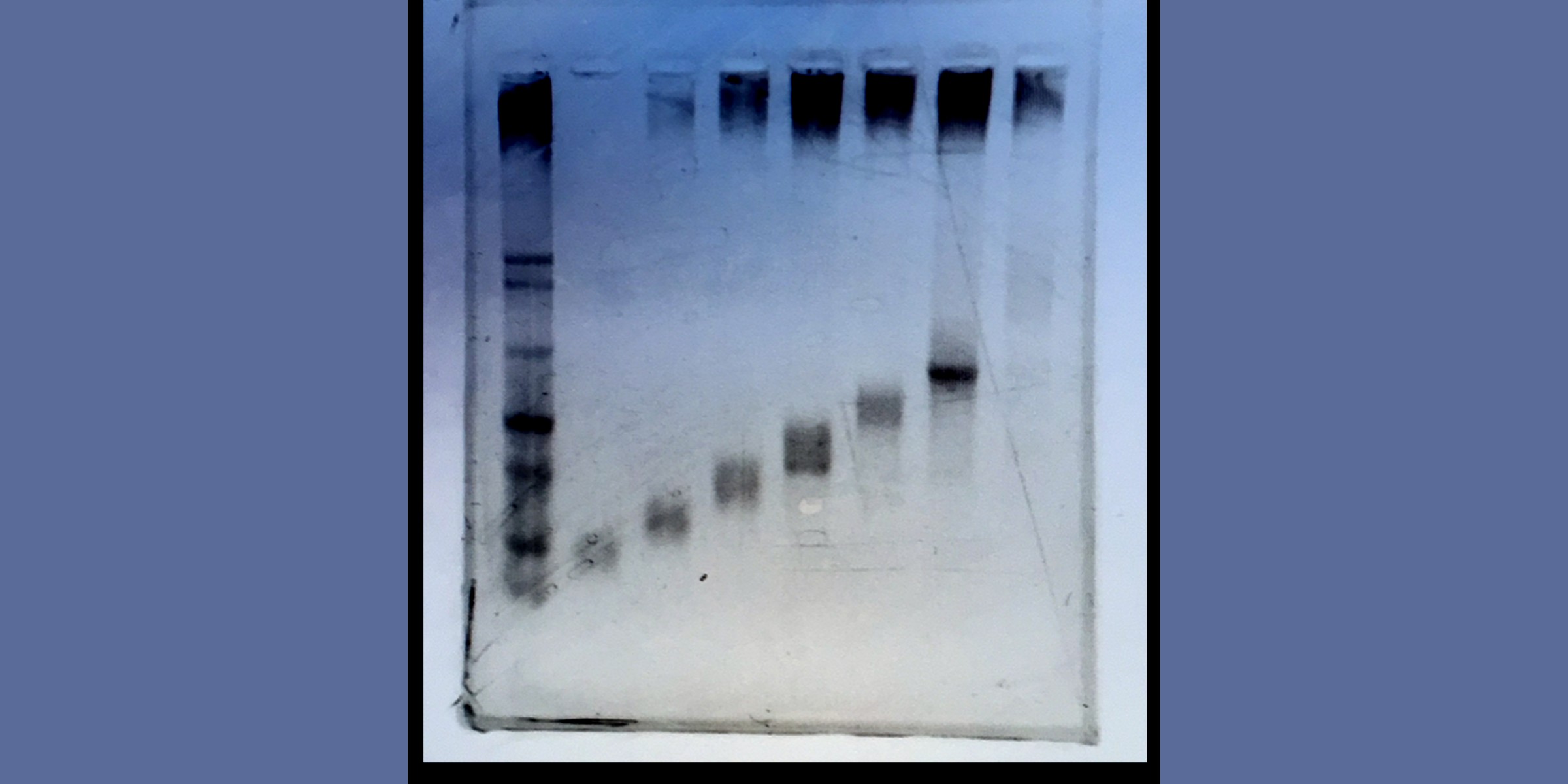
a, Ladder-seq uses a denaturing agarose gel to separate mRNA by length into discrete bands before library preparation and sequencing. Each band contains transcripts of a certain length range that depends on the location of cuts through the gel. The originating band of the resulting reads is tracked using barcodes. In our dataset of mouse NPCs, Ladder-seq reveals transcript Paip2b-204 that contains intronic sequence of transcript Paip2b-201. b, Assessment of length separation by denaturing gel electrophoresis. Length-separated mRNA was run on a new denaturing agarose gel with each band loaded into a separate lane. This assay was conducted once. c, In silico gel. For every annotated transcript, the intensity of a point with y coordinate equal to its annotated length (plus 200-nt average poly(A) tail size) shows the fraction of reads obtained from each band (x axis) that can be assigned unambiguously to it.
Sample application in neurobiology
Using a sample application, the LMU researchers have demonstrated what Ladder-seq can accomplish. In the experiment, they looked at alternative splicing in the developing mouse brain. If you remove a chemical modification of RNA, the sequence of the RNA molecule itself changes. That is to say, you get alternative splicing. From the same gene, a different mRNA is produced — and consequently a different protein, which lacks certain structural domains. They no longer fulfill their biological roles in the brains of mice. Neural stem cells remain in this stage of development. And they no longer differentiate into nerve cells, because corresponding genes are dysregulated. The brains of affected rodents are strongly deformed and the creatures die shortly after birth.
“Without the new method, we would never have been able to answer such questions with such precision and detail,” explains Canzar.
RNA-molecules in the electric field
Source – Ludwig-Maximilians-Universitaet Muenchen (LMU)
05:00 AM
Long read sequencing reveals novel isoforms and insights into splicing regulation during cell state changes [RNA-Seq Blog] 05:00 AM, Wednesday, 12 January 2022 01:20 PM, Wednesday, 12 January 2022
Alternative splicing is a key mechanism underlying cellular differentiation and a driver of complexity in mammalian neuronal tissues. However, understanding of which isoforms are differentially used or expressed and how this affects cellular differentiation remains unclear. Long read sequencing allows full-length transcript recovery and quantification, enabling transcript-level analysis of alternative splicing processes and how these change with cell state. Researchers at the Earlham Institute utilise Oxford Nanopore Technologies sequencing to produce a custom annotation of a well-studied human neuroblastoma cell line SH-SY5Y, and to characterise isoform expression and usage across differentiation.
The researchers identify many previously unannotated features, including a novel transcript of the voltage-gated calcium channel subunit gene, CACNA2D2. They show differential expression and usage of transcripts during differentiation identifying candidates for future research into state change regulation.
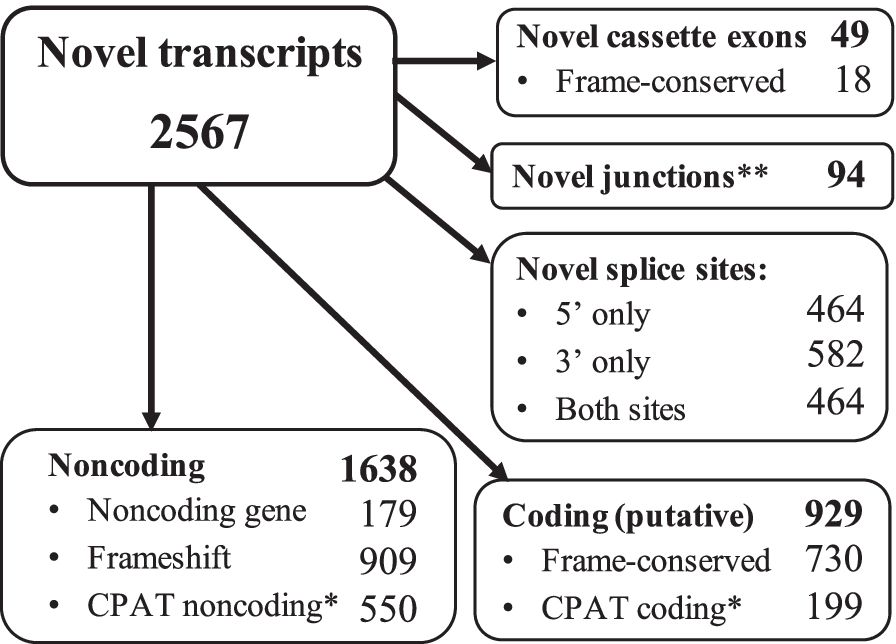
Breakdown of novel transcripts identified using ONT long reads and TALON custom transcriptome annotation
Cassette exons are previously unannotated positions. *CPAT assessment of CDS coding probability (CP ≥ 0.364). **Novel junctions are previously unannotated junctions identified between existing exonic parts. All novel assessments are relative to Gencode v29 human transcriptome annotation
This work highlights the potential of long read sequencing to uncover previously unknown transcript diversity and mechanisms influencing alternative splicing.
04:00 AM
Lucence’s LiquidHALLMARK Sequencing-based Liquid Biopsy Assay Expands to Include cfRNA [RNA-Seq Blog] 04:00 AM, Wednesday, 12 January 2022 01:20 PM, Wednesday, 12 January 2022

Lucence, a precision oncology company using liquid biopsy to bring clarity to cancer care, is announcing the availability of an expanded version of its flagship LiquidHALLMARK liquid biopsy assay that includes both cell-free DNA (cfDNA) and cell-free RNA (cfRNA) profiling. LiquidHALLMARK cfDNA and cfRNA is currently available to US oncologists as a laboratory developed test and is being utilized by physicians at NCI-designated centers across the country.
Most liquid biopsies available today interrogate cell-free DNA (cfDNA) as the sole analyte. However, for the detection of certain fusions, including those that have long intronic regions such as NTRK and NRG1, incorporation of RNA sequencing can be advantageous. Gene fusions like NTRK and NRG1 have emerged as clinically significant and actionable biomarkers that can help guide physicians to matched targeted therapies. Ryma Benayed and co-authors demonstrated that adding targeted RNA sequencing to DNA sequencing to tissue testing could obtain an additional 14% yield of clinically actionable alterations.
An internal validation study conducted by Lucence also supports this approach. Out of 112 non-small cell lung cancer (NSCLC) samples, 29 fusions were detected with a combined cfDNA and cfRNA approach in plasma, compared with 20 fusions from cfDNA alone.
“Including cfRNA in a liquid biopsy assay has a significant impact on the detection of clinically relevant fusions. This solves a major known limitation of cfDNA-only assays, which have more limited fusion detection capabilities. For example, NTRK fusions which have corresponding approved TRK inhibitors, and NRG fusions which have multiple emerging therapies, are best detected by a combination of both DNA and RNA testing. This is an exciting opportunity to help more cancer patients through broader and more accurate targeting,” said Professor Gilberto Lopes, Chief of Medical Oncology at Sylvester Cancer Center at the University of Miami.
“Lucence’s experience using AmpliMark for SARS-CoV-2 as part of the fight against the COVID-19 pandemic gave us the experience with RNA to be able to expand our assay to detect fusions in cancer. It’s a great example of how R&D developed to fight COVID can now be used for ultrasensitive cancer diagnostics,” said Dr. Min-Han Tan, MBBS, PhD, Founding CEO and Medical Director of Lucence.
Lucence’s LiquidHALLMARK cfDNA and cfRNA panel combines cfRNA profiling of 27 actionable and emerging fusions with cfDNA profiling of mutations in 80 genes, fusions in 10 genes, and somatic variants in 15 cancer types. LiquidHALLMARK is powered by AmpliMark, the Company’s proprietary amplicon-based sequencing technology, which uses a unique molecular barcode and error-correction technology to ensure test sensitivity across multiple mutation types for single nucleotide variants and fusion genes.
Source – BusinessWire